



===============
Y Haplogroup R1b
From Wikipedia, the free encyclopedia
(Redirected from R1b)
Haplogroup R1b
 |
|
| Possible time of origin | less than 18,500 years BP[1] |
| Possible place of origin | West Asia[2] |
| Ancestor | R1 |
| Descendants | R1b1a (R-P297), R1b1b (R-M335), R1b1c (R-V88) |
|---|---|
| Defining mutations | 1. M343 defines R1b in the broadest sense 2. P25 defines R1b1, making up most of R1b, and is often used to test for R1b 3. In some cases, major downstream mutations such as M269 are used to identify R1b, especially in regional or out-of-date studies |
| Highest frequencies | British Isles, Western Europe, Northern Cameroon, Bashkirs from Abzelilovsky District |
Contents |
Nomenclature
"R1b", "R1b1", and so on are "phylogenetic" or family tree based names which explain the branching of the family tree of R1b. For example R1b1a and R1b1b would be branches of R1b1, descending from a common ancestor. This means that these names can change with new discoveries.The alternative way of naming haplogroups is to refer to the SNP mutations used to define and identify them, for example "R-M343" which is equivalent to "R1b." Haplogroup R1b is in other words now identified by the presence of the single-nucleotide polymorphism (SNP) mutation M343, which was discovered in 2004.[3] From 2002 to 2005, R1b was defined by the presence of the SNP named P25.
Standardized naming as described above, both using phylogenetic or mutational systems, was first proposed in 2002 by the Y Chromosome Consortium. Prior to 2002, today's Haplogroup R1b had a number of names in differing nomenclature systems, such as Hg1 and Eu18.[4]
Further information: Conversion table for Y chromosome haplogroups
After 2002, a major update of the YCC phylogenetic nomenclature was
made in 2008 by Karafet et al. which took account of newer discoveries
of branches which could be clearly defined by SNP mutations, including
some which changed the understanding of R1b's family tree.[1] Since 2008 it has become increasing necessary to refer to the frequently updated listing made on the ISOGG website.[2]Also prior to 2002, major Y DNA signatures based on markers other than SNPs were recognized. In Western Europe the STR haplotype known as the Atlantic Modal Haplotype was found to be most common by Wilson et al.[5] Even earlier research using RFLP genotyping identified two distinct haplotypes within R-M269. In southeast Europe and southwest Asia (e.g. the Balkans, Georgia and Turkey) "haplotype 35" or "ht35" was found to be a common form of R-M269, whereas in western Europe "haplotype 15" or "ht15" dominated in frequency.[3]
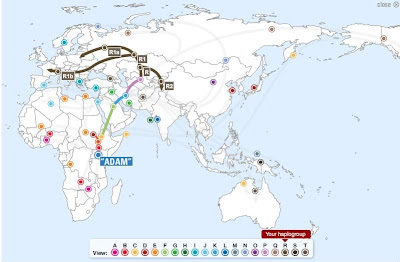
Origin and dispersal
R1b is a sub-clade within the much larger Eurasian MNOPS "macro-haplogroup", which is one of the predominant groupings of all the rest of human male lines outside of Africa, and this whole group, along indeed with all of macro-haplogroup F, is believed to have originated in Asia.| Macro-haplogroup MNOPS |
|
||||||||||||||||||||||||||||||||||||
Early research focused upon Europe. In 2000 Ornella Semino and colleagues argued that R1b had been in Europe before the end of Ice Age, and had spread north from an Iberian refuge after the Last Glacial Maximum.[7] Age estimates of R1b in Europe have steadily decreased in more recent studies, at least concerning the majority of R1b, with more recent studies suggesting a Neolithic age or younger.[6][8][9][10] Only Morelli et al. have recently attempted to defend a Palaeolithic origin for R1b1b2.[11] Irrespective of STR coalescence calculations, Chikhi et al. pointed out that the timing of molecular divergences does not coincide with population splits; the TMRCA of haplogroup R1b (whether in the Palaeolithic or Neolithic) dates to its point of origin somewhere in Eurasia, and not its arrival in western Europe.[1]
Barbara Arredi and colleagues were the first to point out that the distribution of R1b STR variance in Europe forms a cline from east to west, which is more consistent with an entry into Europe from Western Asia with the spread of farming.[10] A 2009 paper by Chiaroni et al. added to this perspective by using R1b as an example of a wave haplogroup distribution, in this case from east to west.[12] The proposal of a southeastern origin of R1b were supported by three detailed studies based on large datasets published in 2010. These detected that the earliest subclades of R1b are found in western Asia and the most recent in western Europe.[6][8][13] While age estimates in these articles are all more recent than the Last Glacial Maximum, all mention the Neolithic, when farming was introduced to Europe from the Middle East as a possible candidate period. Myres et al. (August 2010), and Cruciani et al. (August 2010) both remained undecided on the exact dating of the migration or migrations responsible for this distribution, not ruling out migrations as early as the Mesolithic or as late as Hallstatt but more probably Late Neolithic.[6] They noted that direct evidence from ancient DNA may be needed to resolve these gene flows.[6] Lee et al. (May 2012) analysed the ancient DNA of human remains from the Late Neolithic Bell Beaker site of Kromsdorf, Germany identifying two males as belonging to the Y haplogroup R1b.[14] Analysis of ancient Y DNA from the remains of populations derived from early Neolithic settlements such as the Mediterranean Cardium and Central and North European LBK settlements have found an absence of males belonging to haplogroup R1b.[15][16]
European R1b is now known to be dominated by R-M269, and the origins of this branch are discussed further in more detail below.
Root of R1b tree
For clarity, the identifiers below are those from both the 2010 and 2011 revisions of the ISOGG tree.[2]| 2010 ISOGG tree | 2011 ISOGG tree | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
R1b (R-M343)
R1b* (that is R1b with no subsequent distinguishing SNP mutations) is extremely rare. The only population yet recorded with a definite significant proportion of R1b* are the Kurds of southeastern Kazakhstan with 13%.[6] However, more recently, a large study of Y-chromosome variation in Iran, revealed R1b* as high as 4.3% among Persian sub-populations.[17] In a study of Jordan it was found that no less than 20 out of all 146 men tested (13.7%), including most notably 20 out of 45 men tested from the Dead Sea area, were positive for M173 (R1) but negative for P25 and M269, mentioned above, as well as the R1a markers SRY10831.2 and M17, so they are either R1b* or R1a*.[18] Hassan et al. (2008) found an equally surprising 14 out of 26 (54%) of Sudanese Fulani who were M173+ and P25-.[19] Wood et al. report 2 Egyptian cases of R1-M173 which were negative for SRY10831 (R1a1) and P25 (R1b1), out of a sample of 1,122 males from various African countries, including 92 from Egypt.[20] Such cases could possibly be either R1b* (R-M343*) or R1a* (R-M420*) (demonstrating the importance of checking exact mutations tested when comparing findings in this field).It is however also possible that some of the rare examples represent a reversion of marker P25 from a positive back to a negative ancestral state.[21]
Frequency Table of R1b1 (R-P25) Subclades
An up-to-date compilation of data taking the latest information into account can be found in Cruciani et al. (2010) which can be summarised as follows.[22] As will be discussed below however, in some parts of western and northwestern Europe, R-M269 frequencies can reach even higher levels.| Continent | Population | #No. | Total% | R-P25* | R-V88 | R-M269 | R-M73 |
| Africa | Northern Africa | 691 | 5.9% | 0.0% | 5.2% | 0.7% | 0.0% |
| Africa | Central Sahel Region | 461 | 23.0% | 0.0% | 23.0% | 0.0% | 0.0% |
| Africa | Western Africa | 123 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Eastern Africa | 442 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Southern Africa | 105 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Europe | Western Europeans | 465 | 57.8% | 0.0% | 0.0% | 57.8% | 0.0% |
| Europe | North western Europeans | 43 | 55.8% | 0.0% | 0.0% | 55.8% | 0.0% |
| Europe | Central Europeans | 77 | 42.9% | 0.0% | 0.0% | 42.9% | 0.0% |
| Europe | North Eastern Europeans | 74 | 1.4% | 0.0% | 0.0% | 1.4% | 0.0% |
| Europe | Russians | 60 | 6.7% | 0.0% | 0.0% | 6.7% | 0.0% |
| Europe | Eastern Europeans | 149 | 20.8% | 0.0% | 0.0% | 20.8% | 0.0% |
| Europe | Balkanians | 510 | 13.1% | 0.0% | 0.2% | 12.9% | 0.0% |
| Asia | Western Asians | 328 | 5.8% | 0.0% | 0.3% | 5.5% | 0.0% |
| Asia | Southern Asians | 288 | 4.8% | 0.0% | 0.0% | 1.7% | 3.1% |
| Asia | South eastern Asians | 10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | North eastern Asians | 30 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | Eastern Asians | 156 | 0.6% | 0.0% | 0.0% | 0.6% | 0.0% |
| TOTAL | 5326 |
R1b1 (R-P25)
R1b1*, like R1b* is rare. As mentioned above, examples are described in older articles, for example two in a sample from Turkey,[3] but most cases, especially in Africa, are now thought to be almost mostly in the more recently discovered sub-clade R-V88 (see below). Most or all examples of R1b therefore fall into subclades R1b1a (R-V88) or R1b1b (R-P297). Cruciani et al. in the large 2010 study found 3 cases amongst 1173 Italians, 1 out of 328 West Asians and 1 out of 156 East Asians.[22] Varzari found 3 cases in the Ukraine, in a study of 322 people from the Dniester-Carpathian region, who were P25 positive, but M269 negative.[23] Cases from older studies are mainly from Africa, the Middle East or Mediterranean, and are discussed below as probable cases of R1b1a (R-V88).R1b1a (R-P297)
R1b1a is defined by the presence of SNP marker P297. In 2008 this polymorphism was recognised to combine M73 and M269 into one R1b1a cluster.[1] The majority of Eurasian R1b is within this clade, representing a very large modern population. Although P297 itself has not yet been much tested for, the same population has been relatively well studied in terms of other markers. Therefore the branching within this clade can be explained in relatively high detail below.R1b1a1 (R-M73)
R1b1a1 (2011 name) is defined by the presence of SNP marker M73. It has been found at generally low frequencies throughout central Eurasia,[24] but has been found with relatively high frequency among particular populations there including Hazaras in Pakistan (8/25 = 32%);[25] and Bashkirs in Bashkortostan (62/471 = 13.2%), 44 of these being found among the 80 tested Bashkirs of the Abzelilovsky District in the Republic of Bashkortostan (55.0%).[26] Four R-M73 men were also found in a 523-person study of Turkey,[3] and one person in a 168-person study of Crete.[27]In 2010, Myres et al. report that out of 193 R-M73 men found amongst 10,355 widespread men, "all except two Russians occurred outside Europe, either in the Caucasus, Turkey, the Circum-Uralic and North Pakistan regions."[28]
R1b1a2 (R-M269)
R1b1a2 (2011 name) is defined by the presence of SNP marker M269. R1b1a2* or M269(xL23) is found at highest frequency in the central Balkans notably Kosovo with 7.9%, Macedonia 5.1% and Serbia 4.4%.[6] Kosovo is notable in also having a high percentage of descendant L23* or L23(xM412) at 11.4% unlike most other areas with significant percentages of M269* and L23* except for Poland with 2.4% and 9.5% and the Bashkirs of southeast Bashkortostan with 2.4% and 32.2% respectively.[6] Notably this Bashkir population also has a high percentage of M269 sister branch M73 at 23.4%.[6] Five individuals out of 110 tested in the Ararat Valley, Armenia belonged to R1b1a2* and 36 to L23*, with none belonging to subclades of L23.[29]European R1b is dominated by R-M269. It has been found at generally low frequencies throughout central Eurasia,[24] but with relatively high frequency among Bashkirs of the Perm Region (84.0%).[26] This marker is also present in China and India at frequencies of less than one percent. The table below lists in more detail the frequencies of M269 in various regions in Asia, Europe, and Africa.
The frequency is about 71% in Scotland, 70% in Spain and 60% in France. In south-eastern England the frequency of this clade is about 70%; in parts of the rest of north and western England, Spain, Portugal, Wales and Ireland, it is as high as 90%; and in parts of north-western Ireland it reaches 98%. It is also found in North Africa, where its frequency surpasses 10% in some parts of Algeria.[30]
From 2003 to 2005 what is now R1b1a2 was designated R1b3. From 2005 to 2008 it was R1b1c. From 2008 to 2011 it was R1b1b2.
| M269 |
|
||||||||||||||||||||||||||||||||||||
It was also in this period between 2000 and 2010 that it became clear that especially Western European R1b is dominated by specific sub-clades of R-M269 (with some small amounts of other types found in areas such as Sardinia[6][11]). Within Europe, R-M269 is dominated by R-M412, also known as R-L51, which according to Myres et al. (2010) is "virtually absent in the Near East, the Caucasus and West Asia." This Western European population is further divided between R-P312/S116 and R-U106/S21, which appear to spread from the western and eastern Rhine river basin respectively. Myres et al. note further that concerning its closest relatives, in R-L23*, that it is "instructive" that these are often more than 10% of the population in the Caucasus, Turkey, and some southeast European and circum-Uralic populations. In Western Europe it is also present but in generally much lower levels apart from "an instance of 27% in Switzerland's Upper Rhone Valley."[6] In addition, the sub-clade distribution map, Figure 1h titled "L11(xU106,S116)", in Myres et al. shows that R-P310/L11* (or as yet undefined subclades of R-P310/L11) occurs only in frequencies greater than 10% in Central England with surrounding areas of England and Wales having lower frequencies.[6] This R-P310/L11* is almost non-existent in the rest of Eurasia and North Africa with the exception of coastal lands fringing the western and southern Baltic (reaching 10% in Eastern Denmark and 6% in northern Poland) and in Eastern Switzerland and surrounds.[6]
In 2009, DNA extracted from the femur bones of 6 skeletons in an early-medieval burial place in Ergolding (Bavaria, Germany) dated to around 670 AD yielded the following results: 4 were found to be haplogroup R1b with the closest matches in modern populations of Germany, Ireland and the USA while 2 were in Haplogroup G2a.[32]
Population studies which test for M269 have become more common in recent years, while in earlier studies men in this haplogroup are only visible in the data by extrapolation of what is likely. The following gives a summary of most of the studies which specifically tested for M269, showing its distribution in Europe, North Africa, the Middle East and Central Asia as far as China and Nepal.
| Country | Sampling | sample | R-M269 | Source |
| Wales | National | 65 | 92.3% | Balaresque et al. (2009)[8] |
| Spain | Basques | 116 | 87.1% | Balaresque et al. (2009)[8] |
| Ireland | National | 796 | 85.4% | Moore et al. (2006)[33] |
| Spain | Catalonia | 80 | 81.3% | Balaresque et al. (2009)[8] |
| France | Ille-et-Vilaine | 82 | 80.5% | Balaresque et al. (2009)[8] |
| France | Haute-Garonne | 57 | 78.9% | Balaresque et al. (2009)[8] |
| England | Cornwall | 64 | 78.1% | Balaresque et al. (2009)[8] |
| France | Loire-Atlantique | 48 | 77.1% | Balaresque et al. (2009)[8] |
| France | Finistère | 75 | 76.0% | Balaresque et al. (2009)[8] |
| France | Basques | 61 | 75.4% | Balaresque et al. (2009)[8] |
| Spain | East Andalucia | 95 | 72.0% | Balaresque et al. (2009)[8] |
| Spain | Castilla La Mancha | 63 | 72.0% | Balaresque et al. (2009)[8] |
| France | Vendée | 50 | 68.0% | Balaresque et al. (2009)[8] |
| France | Baie de Somme | 43 | 62.8% | Balaresque et al. (2009)[8] |
| England | Leicestershire | 43 | 62.0% | Balaresque et al. (2009)[8] |
| Italy | North-East (Ladin) | 79 | 60.8% | Balaresque et al. (2009)[8] |
| Spain | Galicia | 88 | 58.0% | Balaresque et al. (2009)[8] |
| Spain | West Andalucia | 72 | 55.0% | Balaresque et al. (2009)[8] |
| Portugal | South | 78 | 46.2% | Balaresque et al. (2009)[8] |
| Italy | North-West | 99 | 45.0% | Balaresque et al. (2009)[8] |
| Denmark | National | 56 | 42.9% | Balaresque et al. (2009)[8] |
| Netherlands | National | 84 | 42.0% | Balaresque et al. (2009)[8] |
| Italy | North East | 67 | 41.8% | Battaglia et al. (2008)[34] |
| Russia | Bashkirs | 471 | 34.40% | Lobov (2009)[26] |
| Germany | Bavaria | 80 | 32.3% | Balaresque et al. (2009)[8] |
| Italy | West Sicily | 122 | 30.3% | Di Gaetano et al. (2009)[35] |
| Poland | National | 110 | 22.7% | Myres et al. (2007)[28] |
| Slovenia | National | 75 | 21.3% | Battaglia et al. (2008)[34] |
| Slovenia | National | 70 | 20.6% | Balaresque et al. (2009)[8] |
| Turkey | Central | 152 | 19.1% | Cinnioğlu et al. (2004)[3] |
| Republic of Macedonia | National | 64 | 18.8% | Battaglia et al. (2008)[34] |
| Italy | East Sicily | 114 | 18.4% | Di Gaetano et al. (2009)[35] |
| Crete | National | 193 | 17.0% | King et al. (2008)[36] |
| Italy | Sardinia | 930 | 17.0% | Contu et al. (2008)[37] |
| Iran | North | 33 | 15.2% | Regueiro et al. (2006)[38] |
| Moldova | 268 | 14.6% | Varzari (2006)[23] | |
| Greece | National | 171 | 13.5% | King et al. (2008)[36] |
| Turkey | West | 163 | 13.5% | Cinnioğlu et al. (2004)[3] |
| Romania | National | 54 | 13.0% | Varzari (2006)[23] |
| Turkey | East | 208 | 12.0% | Cinnioğlu et al. (2004)[3] |
| Algeria | Northwest (Oran area) | 102 | 11.8% | Robino et al. (2008)[39] |
| Russia | Roslavl | 107 | 11.2% | Balanovsky et al. (2008)[40] |
| Iraq | National | 139 | 10.8% | Al-Zahery et al. (2003)[41] |
| Nepal | Newar | 66 | 10.60% | Gayden et al. (2007)[42] |
| Serbia | National | 100 | 10.0% | Belaresque et al. (2009)[8] |
| Tunisia | Tunis | 139 | 7.2% | Adams et al. (2008)[43] |
| Algeria | Algiers, Tizi Ouzou | 46 | 6.5% | Adams et al. (2008)[43] |
| Bosnia-Herzegovina | Serb | 81 | 6.2% | Marjanovic et al. (2005)[44] |
| Iran | South | 117 | 6.0% | Regueiro et al. (2006)[38] |
| Russia | Repievka | 96 | 5.2% | Balanovsky et al. (2008)[40] |
| UAE | 164 | 3.7% | Cadenas et al. (2007)[45] | |
| Bosnia-Herzegovina | Bosniak | 85 | 3.5% | Marjanovic et al. (2005)[44] |
| Pakistan | 176 | 2.8% | Sengupta et al. (2006)[25] | |
| Russia | Belgorod | 143 | 2.8% | Balanovsky et al. (2008)[40] |
| Russia | Ostrov | 75 | 2.7% | Balanovsky et al. (2008)[40] |
| Russia | Pristen | 45 | 2.2% | Balanovsky et al. (2008)[40] |
| Bosnia-Herzegovina | Croat | 90 | 2.2% | Marjanovic et al. (2005)[44] |
| Qatar | 72 | 1.4% | Cadenas et al. (2007)[45] | |
| China | 128 | 0.8% | Sengupta et al. (2006)[25] | |
| India | various | 728 | 0.5% | Sengupta et al. (2006)[25] |
| Croatia | Osijek | 29 | 0.0% | Battaglia et al. (2008)[34] |
| Yemen | 62 | 0.0% | Cadenas et al. (2007)[45] | |
| Tibet | 156 | 0.0% | Gayden et al. (2007)[42] | |
| Nepal | Tamang | 45 | 0.0% | Gayden et al. (2007)[42] |
| Nepal | Kathmandu | 77 | 0.0% | Gayden et al. (2007)[42] |
| Japan | 23 | 0.0% | Sengupta et al. (2006)[25] |
R1b1a2a1a1a (R-U106)
This subclade is defined by the presence of the SNP U106, also known as S21 and M405.[2][46] It appears to represent over 25% of R1b in Europe and is most common in the Netherlands and the northern half of Belgium.[2]| U106/S21 |
|
|||||||||||||||||||||||||||||||||||||||
| Population | Sample size | R-M269 | R-U106 | R-U106-1 |
| Austria [28] | 22 | 27% | 23% | 0.0% |
| Central/South America [28] | 33 | 0.0% | 0.0% | 0.0% |
| Czech Republic [28] | 36 | 28% | 14% | 0.0% |
| Denmark [28] | 113 | 34% | 17% | 0.9% |
| Eastern Europe[28] | 44 | 5% | 0.0% | 0.0% |
| England[28] | 138 | 57% | 20% | 1.4% |
| France[28] | 56 | 52% | 7% | 0.0% |
| Germany[28] | 332 | 43% | 19% | 1.8% |
| Ireland[28] | 102 | 80% | 6% | 0.0% |
| Italy[28] | 284 | 37% | 4% | 0.0% |
| Jordan[28] | 76 | 0.0% | 0.0% | 0.0% |
| Middle-East[28] | 43 | 0.0% | 0.0% | 0.0% |
| Netherlands[28] | 94 | 54% | 35% | 2.1% |
| Oceania[28] | 43 | 0.0% | 0.0% | 0.0% |
| Oman[28] | 29 | 0.0% | 0.0% | 0.0% |
| Pakistan[28] | 177 | 3% | 0.0% | 0.0% |
| Palestine[28] | 47 | 0.0% | 0.0% | 0.0% |
| Poland[28] | 110 | 23% | 8% | 0.0% |
| Russia[28] | 56 | 21% | 5.4% | 1.8% |
| Slovenia[28] | 105 | 17% | 4% | 0.0% |
| Spain | 164 | 42% | 8% | 0.0% |
| Switzerland[28] | 90 | 58% | 13% | 0.0% |
| Turkey[28] | 523 | 14% | 0.4% | 0.0% |
| Ukraine[28] | 32 | 25% | 9% | 0.0% |
| United States[28] | 58 | 5% | 5% | 0.0% |
| US (European) | 125 | 46% | 15% | 0.8% |
| US (Afroamerican) | 118 | 14% | 2.5% | 0.8% |
R1b1a2a1a2 (R-P312/S116)
Along with R-U106, R-P312 is one of the most common types of R1b1a2 (R-M269) in Europe. Also known as S116, it has been the subject of significant study concerning its sub-clades, and some of the ones recognized by the ISOGG website are summarized in the following table.[2] Myres et al. described it distributing from the west of the Rhine basin.[6]| P312 |
|


Orgulloso de pertenecer al haplogrupo R1b p312*
ResponderEliminar